A. omnicolens
Emery Haley, PhD, Scientific Writing Specialist
Alloscardovia omnicolens
Clinical Summary
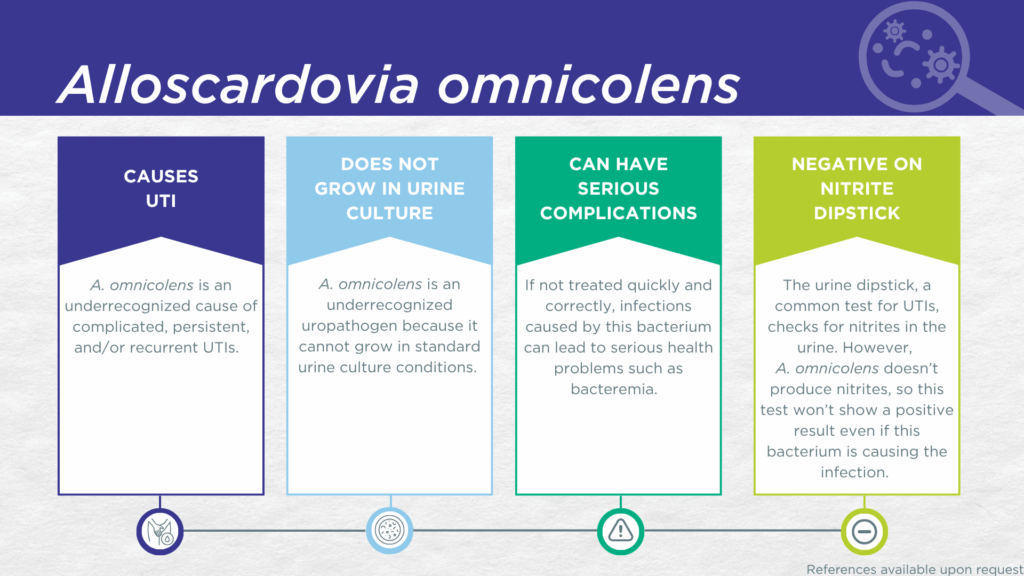
- A. omnicolens is a nitrite-negative, gram-positive, microorganism.
- A. omnicolens is fastidious and cannot grow in standard urine culture conditions.
- A. omnicolens is associated with complicated UTIs in older adults and immune-compromised individuals.
- In symptomatic UTI patients, A. omnicolens:
- Is not a contaminant (is found in catheter-collected urine specimens).
- Is viable (can grow out in expanded culture conditions).
- Is pathogenic (associated with elevated urine biomarkers of infection).
- Reported severe complications of A. omnicolens UTI include bacteremia.
Bacterial Characteristics
Gram-stain
Gram-positive
Morphology
Bacillus
Growth Requirements
Fastidious
Obligate anaerobe
Nitrate Reduction
No
Urease
Negative
Biofilm Formation
No
Pathogenicity
Colonizer or Pathobiont
Clinical Relevance in UTI
A. omnicolens was first identified from clinical specimens, including urine and blood specimens, in 2007.[1]
A. omnicolens UTI is challenging to diagnose. Firstly, A. omnicolens lacks nitrate reductase activity, so screening strategies involving urinalysis for nitrite positivity will be false-negative.[2,3] Secondly, growing A. omnicolens in culture requires an anaerobic atmosphere which is not used in clinical laboratories performing standard culture techniques for UTI diagnosis.[4] Thirdly, even when this organism does grow in culture as a gram-positive bacillus, it is frequently confused with Actinomyces species or labeled as a “coryneform bacterium” and may be dismissed as an irrelevant gram-positive “contaminant” organism from the urogenital microbiome.[5] Instead, A. omnicolens UTI is most often diagnosed by advanced proteomic techniques, such as Matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS), or advanced molecular techniques, including polymerase chain reaction (PCR) and sequencing.[5–7]
A. omnicolens is considered a rare opportunistic UTI pathogen in immune-compromised individuals [4] and has been isolated in pure monoculture at a density of > 105 CFU/mL from patients with lower urinary tract symptoms with or without pyuria on urinalysis.[8] A study using expanded quantitative urine culture (EQUC), a technique growing a larger urine volume with additional nutritional media, different atmospheric conditions, and longer incubations, found that the organism was viable.[9] Additionally, in a study of older adult males and females with clinically suspected complicated UTI, A. omnicolens was detected in both midstream voided and in-and-out-catheter collected specimens indicating that it was truly present in the bladder, not simply a contaminant picked up during voiding.[10] Furthermore, elevated markers of immune system activation in the urinary tract have been measured from the same clinical urine specimens in which A. omnicolens was detected, indicating that the presence of A. omnicolens was associated with an immune response to urinary tract infection.[11–13] Critically, A. omnicolens bacteremia secondary to UTI has been reported.[14]
Together, these findings indicate that A. omnicolens should be seriously considered as a uropathogen and demonstrate the value of detecting this organism, particularly in individuals with immune-compromising comorbidities or other risk factors for complicated UTI.
Treatment
Evidence of Efficacy (Checkmarks) [15]: Amoxicillin/Clavulanate, Ampicillin, Ampicillin/Sulbactam, Cefazolin, Cefepime, Ceftazidime, Ceftriaxone, Cephalexin, Ciprofloxacin, Doxycycline, Linezolid, Piperacillin/Tazobactam, Sulfamethoxazole/Trimethoprim, and Vancomycin.
- Huys, G.; Vancanneyt, M.; D’Haene, K.; Falsen, E.; Wauters, G.; Vandamme, P. Alloscardovia Omnicolens Gen. Nov., Sp. Nov., from Human Clinical Samples. Int J Syst Evol Micr 2007, 57, 1442–1446, doi:10.1099/ijs.0.64812-0.
- BacDive | The Bacterial Diversity Metadatabase Available online: https://bacdive.dsmz.de/ (accessed on 11 February 2025).
- BioCyc Pathway/Genome Database Collection Available online: https://biocyc.org/ (accessed on 11 February 2025).
- Moreland, R.B.; Choi, B.I.; Geaman, W.; Gonzalez, C.; Hochstedler-Kramer, B.R.; John, J.; Kaindl, J.; Kesav, N.; Lamichhane, J.; Lucio, L.; et al. Beyond the Usual Suspects: Emerging Uropathogens in the Microbiome Age. Frontiers in Urology 2023, 3, doi:10.3389/fruro.2023.1212590.
- Lainhart, W.; Gonzalez, M.D. Aerococcus Urinae, Alloscardovia Omnicolens, and Actinotignum Schaalii: The AAA Minor League Team of Urinary Tract Infection Pathogens. Clinical Microbiology Newsletter 2018, 40, 77–82, doi:10.1016/j.clinmicnews.2018.05.001.
- Lainhart, W.; Burnham, C.-A.D. Enhanced Recovery of Fastidious Organisms from Urine Culture in the Setting of Total Laboratory Automation. J Clin Microbiol 2018, 56, doi:10.1128/jcm.00546-18.
- Klein, S.; Nurjadi, D.; Horner, S.; Heeg, K.; Zimmermann, S.; Burckhardt, I. Significant Increase in Cultivation of Gardnerella Vaginalis, Alloscardovia Omnicolens, Actinotignum Schaalii, and Actinomyces Spp. in Urine Samples with Total Laboratory Automation. Eur. J. Clin. Microbiol. Infect. Dis. 2018, 37, 1305–1311, doi:10.1007/s10096-018-3250-6.
- Mahlen, S.D.; Clarridge, J.E. Site and Clinical Significance of Alloscardovia Omnicolens and Bifidobacterium Species Isolated in the Clinical Laboratory▿. J Clin Microbiol 2009, 47, 3289–3293, doi:10.1128/jcm.00555-09.
- Festa, R.A.; Luke, N.; Mathur, M.; Parnell, L.; Wang, D.; Zhao, X.; Magallon, J.; Remedios-Chan, M.; Nguyen, J.; Cho, T.; et al. A Test Combining Multiplex-PCR with Pooled Antibiotic Susceptibility Testing Has High Correlation with Expanded Urine Culture for Detection of Live Bacteria in Urine Samples of Suspected UTI Patients. Diagn Microbiol Infect Dis 2023, 107, 116015, doi:10.1016/j.diagmicrobio.2023.116015.
- Wang, D.; Haley, E.; Luke, N.; Mathur, M.; Festa, R.; Zhao, X.; Anderson, L.A.; Allison, J.L.; Stebbins, K.L.; Diaz, M.J.; et al. Emerging and Fastidious Uropathogens Were Detected by M-PCR with Similar Prevalence and Cell Density in Catheter and Midstream Voided Urine Indicating the Importance of These Microbes in Causing UTIs. Infect. Drug Resist. 2023, Volume 16, 7775–7795, doi:10.2147/idr.s429990.
- Haley, E.; Luke, N.; Mathur, M.; Festa, R.A.; Wang, J.; Jiang, Y.; Anderson, L.A.; Baunoch, D. The Prevalence and Association of Different Uropathogens Detected by M-PCR with Infection-Associated Urine Biomarkers in Urinary Tract Infections. Res. Rep. Urol. 2024, 16, 19–29, doi:10.2147/rru.s443361.
- Akhlaghpour, M.; Haley, E.; Parnell, L.; Luke, N.; Mathur, M.; Festa, R.A.; Percaccio, M.; Magallon, J.; Remedios-Chan, M.; Rosas, A.; et al. Urine Biomarkers Individually and as a Consensus Model Show High Sensitivity and Specificity for Detecting UTIs. BMC Infect Dis 2024, 24, 153, doi:10.1186/s12879-024-09044-2.
- Parnell, L.K.D.; Luke, N.; Mathur, M.; Festa, R.A.; Haley, E.; Wang, J.; Jiang, Y.; Anderson, L.; Baunoch, D. Elevated UTI Biomarkers in Symptomatic Patients with Urine Microbial Densities of 10,000 CFU/ML Indicate a Lower Threshold for Diagnosing UTIs. MDPI 2023, 13, 1–15, doi:10.3390/diagnostics13162688.
- Ogawa, Y.; Koizumi, A.; Kasahara, K.; Lee, S.-T.; Yamada, Y.; Nakano, R.; Yano, H.; Mikasa, K. Bacteremia Secondary to Alloscardovia Omnicolens Urinary Tract Infection. Journal of Infection and Chemotherapy 2016, 22, 424–425, doi:10.1016/j.jiac.2015.12.013.
- Isnard, C.; Lienhard, R.; Reissier, S.; Rodriguez, S.; Krähenbühl, J.; Liassine, N.; Guérin, F.; Cattoir, V. In Vitro Antimicrobial Susceptibility of Alloscardovia Omnicolens and Molecular Mechanisms of Acquired Resistance. Diagnostic Microbiology and Infectious Disease 2016, 84, 227–229, doi:10.1016/j.diagmicrobio.2015.08.009.
Dr. Emery Haley is a scientific writing specialist with over ten years of experience in translational cell and molecular biology. As both a former laboratory scientist and an experienced science communicator, Dr. Haley is passionate about making complex research clear, approachable, and relevant. Their work has been published in over 10 papers and focuses on bridging the gap between the lab and real-world patient care to help drive better health outcomes.